MASH v FLASH GTEx analysis: nonnegative loadings
Last updated: 2018-08-16
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180609)The command
set.seed(20180609)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: be0d4d7
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: data/ Ignored: docs/.DS_Store Ignored: docs/images/.DS_Store Ignored: docs/images/.Rapp.history Ignored: output/.DS_Store Ignored: output/.Rapp.history Ignored: output/MASHvFLASHgtex/.DS_Store Ignored: output/MASHvFLASHsims/.DS_Store Ignored: output/MASHvFLASHsims/backfit/.DS_Store Ignored: output/MASHvFLASHsims/backfit/.Rapp.history
Expand here to see past versions:
Introduction
Here I fit MASH and FLASH objects to the “strong” GTEx dataset using the nonnegative loadings obtained in the previous analysis. See below for code and here for an introduction to the plots below.
Fitting methods
The workflows are similar to those described here.
The workflow for FLASH is identical, except that I use the nonnegative loadings from the previous analysis as my “data-driven” loadings. (To save time, I only use the loadings corresponding to “multi-tissue effects,” since the other loadings are well represented by “canonical” loadings. Further, to aid interpretation, I normalize the loadings so that each has \(\ell_\infty\)-norm equal to 1.)
The workflow for MASH is also identical, except that I use rank-one matrices derived from the nonnegative loadings rather than rank-one matrices obtained using Extreme Deconvolution. More precisely, for each multi-tissue effect \(\ell_i\), I add the matrix \(\ell_i \ell_i^T\) to the list of matrices \(U_i\) in the MASH mixture model.
Results
I pre-run the code and load the fits from file.
library(mashr)Loading required package: ashrdevtools::load_all("/Users/willwerscheid/GitHub/flashr/")Loading flashrsource("./code/gtexanalysis.R")
gtex <- readRDS(gzcon(url("https://github.com/stephenslab/gtexresults/blob/master/data/MatrixEQTLSumStats.Portable.Z.rds?raw=TRUE")))
strong <- t(gtex$strong.z)
fpath <- "./output/MASHvFLASHgtex3/"
m_final <- readRDS(paste0(fpath, "m.rds"))
fl_final <- readRDS(paste0(fpath, "fl.rds"))
m_lfsr <- t(get_lfsr(m_final))
m_pm <- t(get_pm(m_final))
fl_lfsr <- readRDS(paste0(fpath, "fllfsr.rds"))
fl_pm <- flash_get_fitted_values(fl_final)There is significant disagreement between the two fits:
signif_by_mash = as.vector(m_lfsr < 0.05)
signif_by_flash = as.vector(fl_lfsr < 0.05)
round(table(signif_by_mash, signif_by_flash) / length(signif_by_mash),
digits = 2) signif_by_flash
signif_by_mash FALSE TRUE
FALSE 0.31 0.17
TRUE 0.08 0.44Significant for FLASH, not MASH
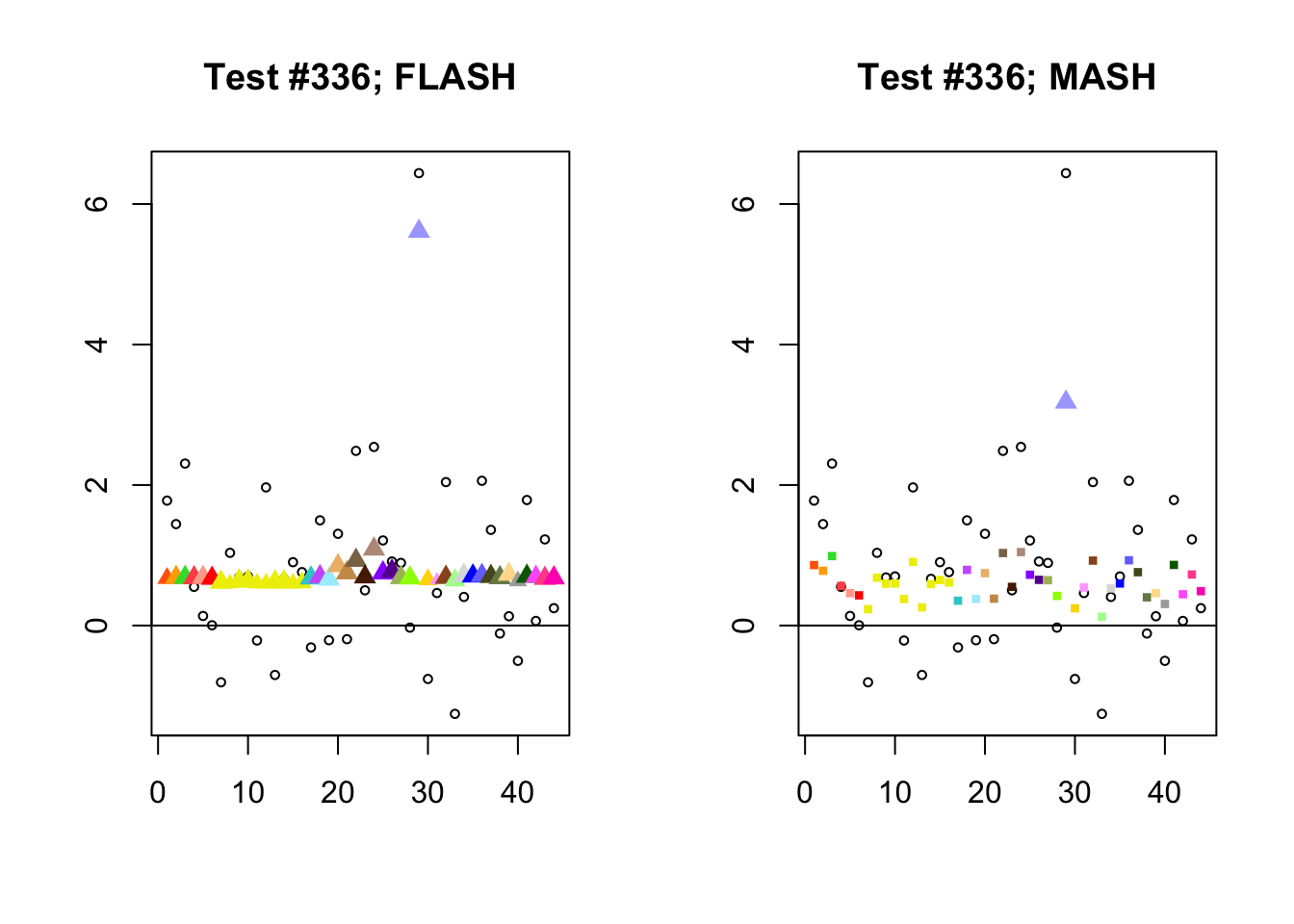
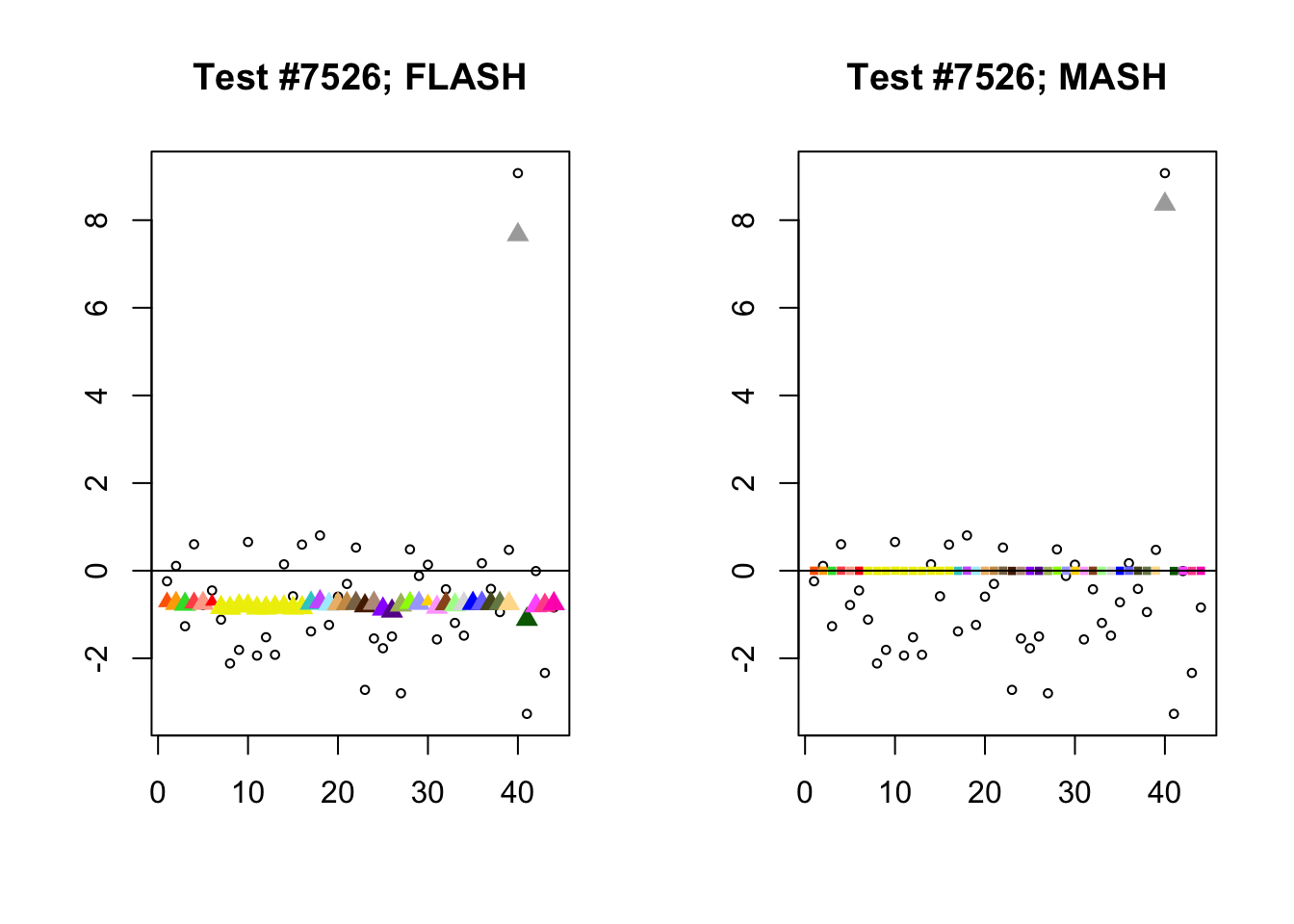
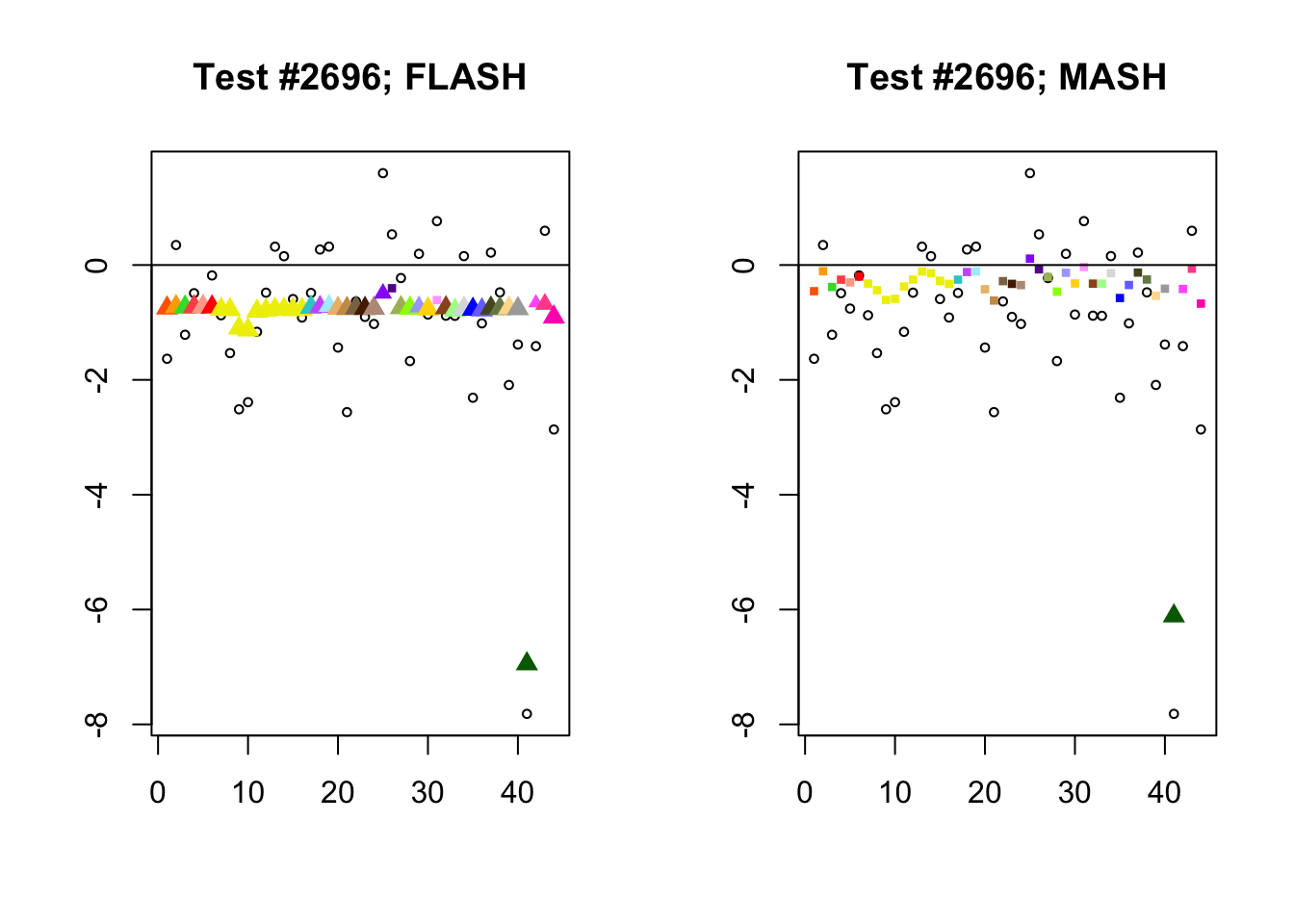
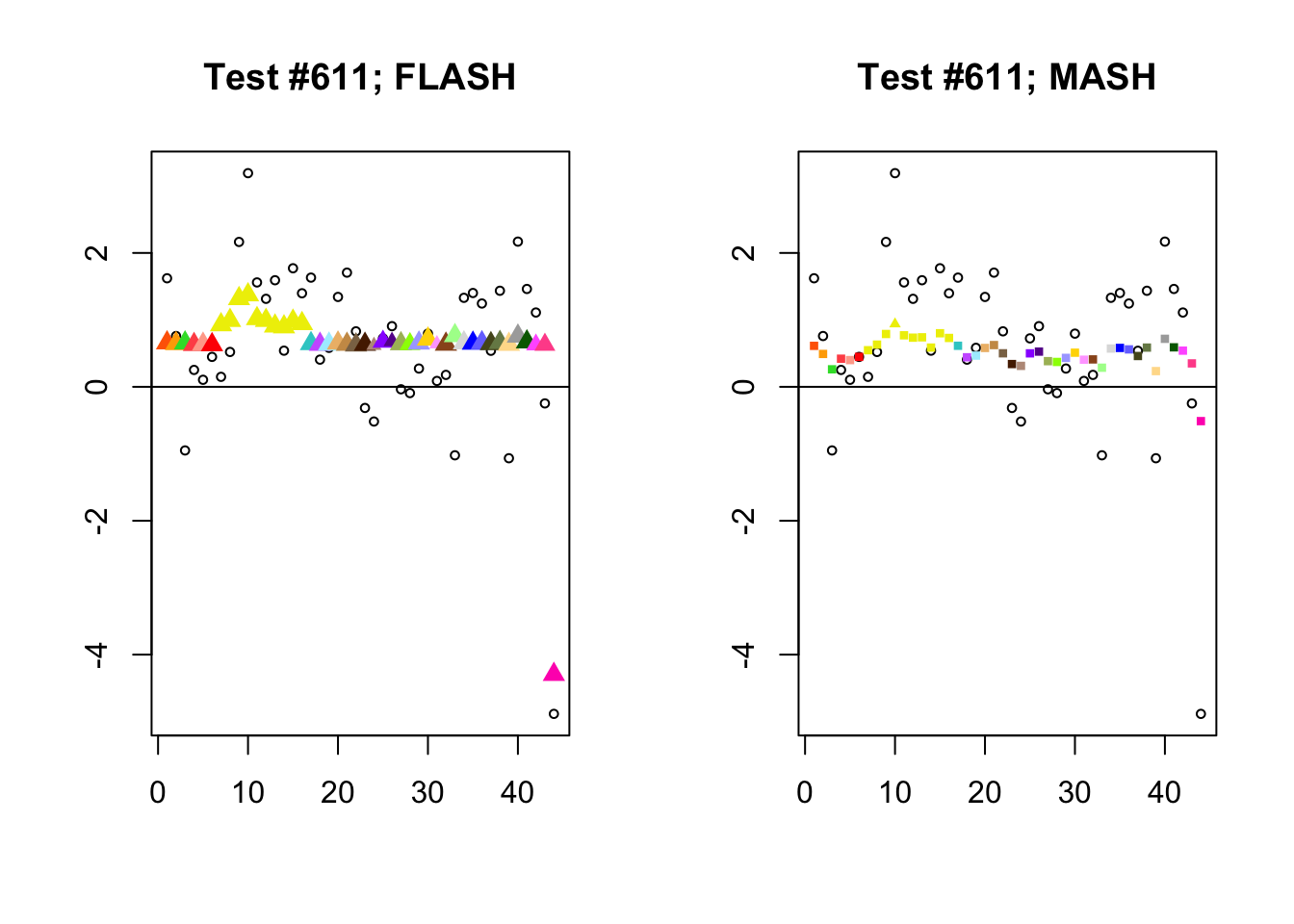
As discussed in previous analyses (see, for example, here), FLASH often finds a combination of a large unique effect and a small equal effect where MASH only finds the unique effect to be significant (or, more rarely, no effects).
# interesting.tests <- compare_methods(fl_lfsr, m_lfsr, fl_pm, m_pm)
par(mfrow=c(1, 2))
identical.plus.unique <- c(336, 7526, 2696, 611)
for (n in identical.plus.unique) {
plot_test(n, fl_lfsr, fl_pm, "FLASH")
plot_test(n, m_lfsr, m_pm, "MASH")
}
Expand here to see past versions of flash_not_mash-1.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of flash_not_mash-2.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of flash_not_mash-3.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of flash_not_mash-4.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
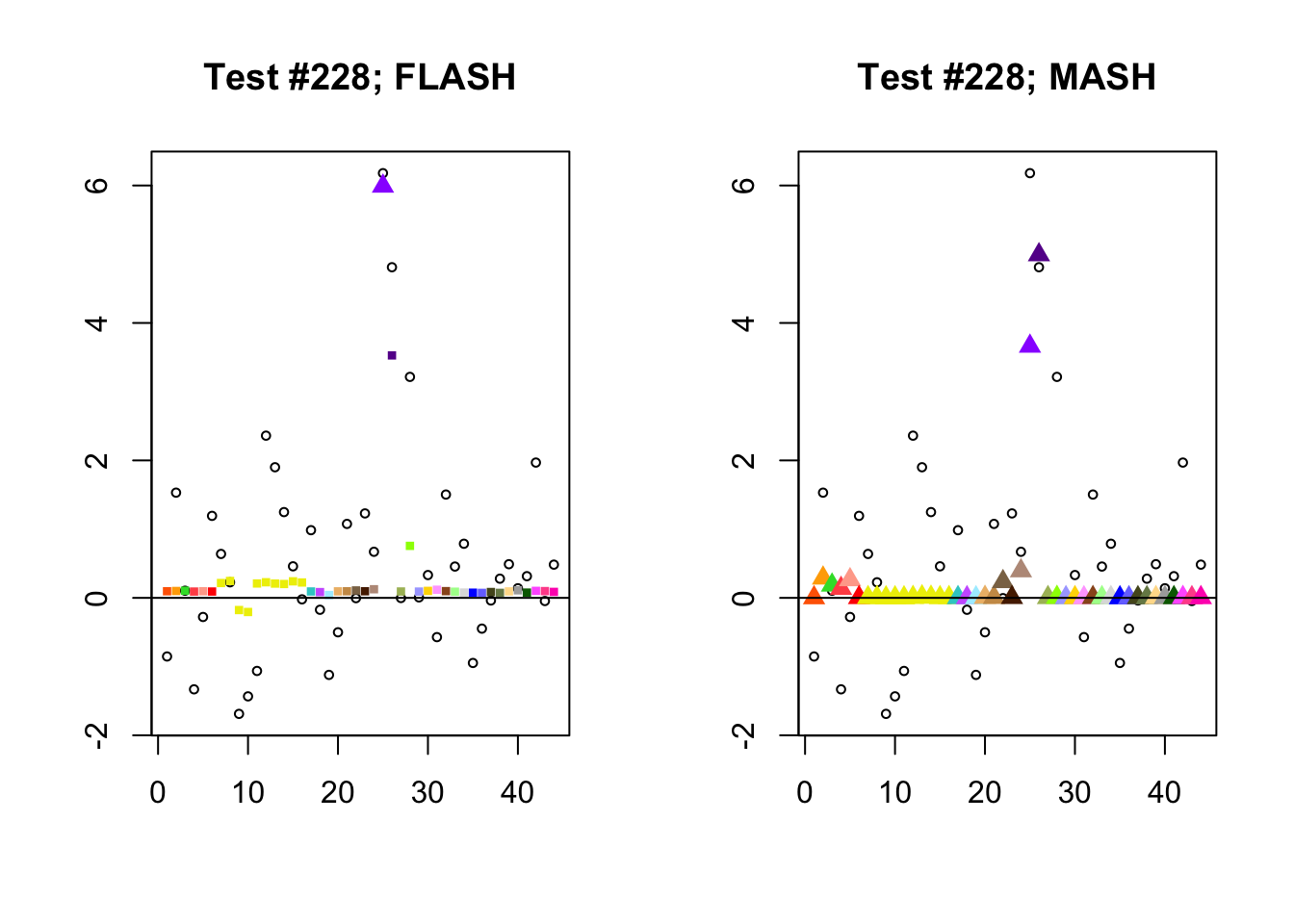
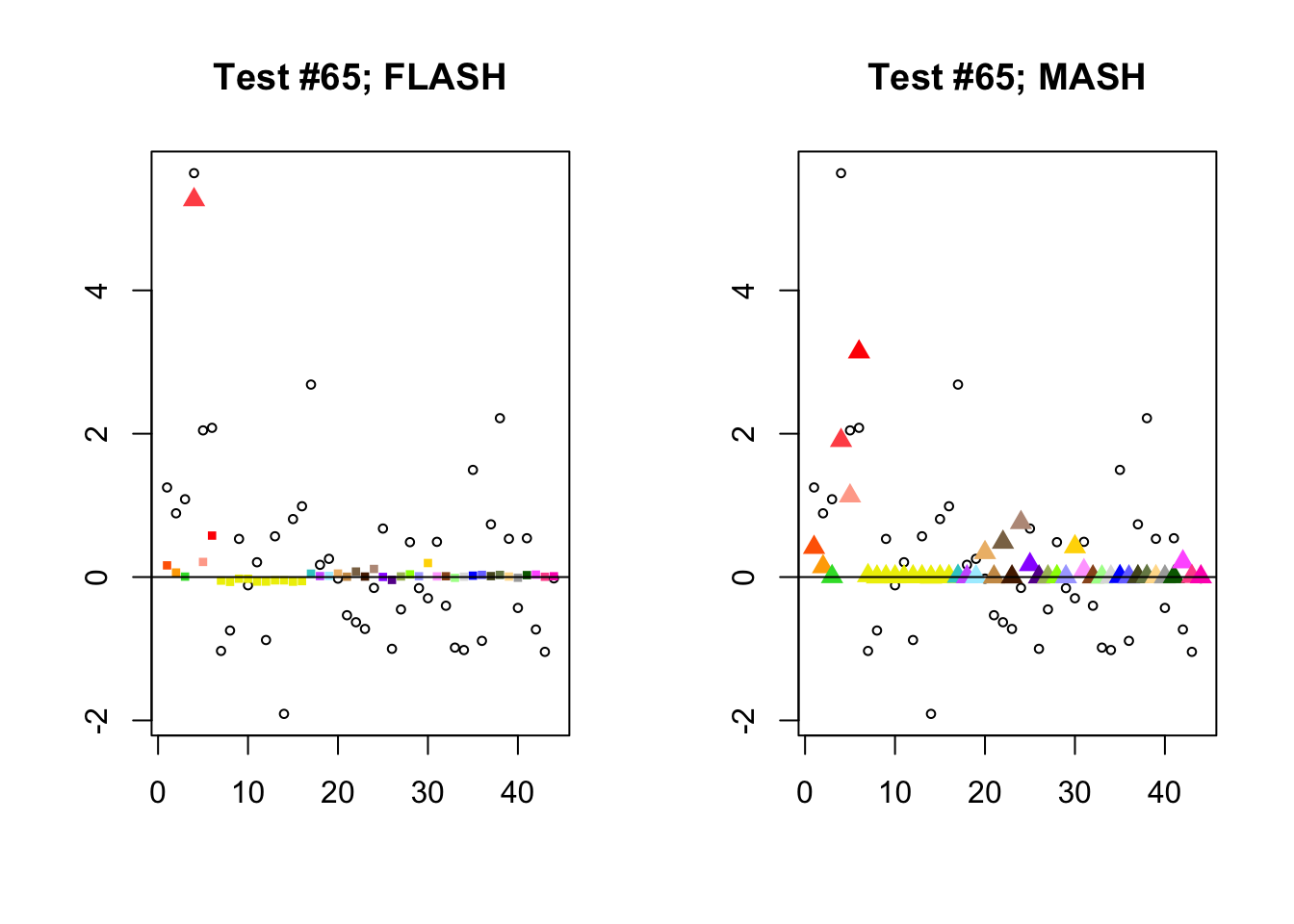
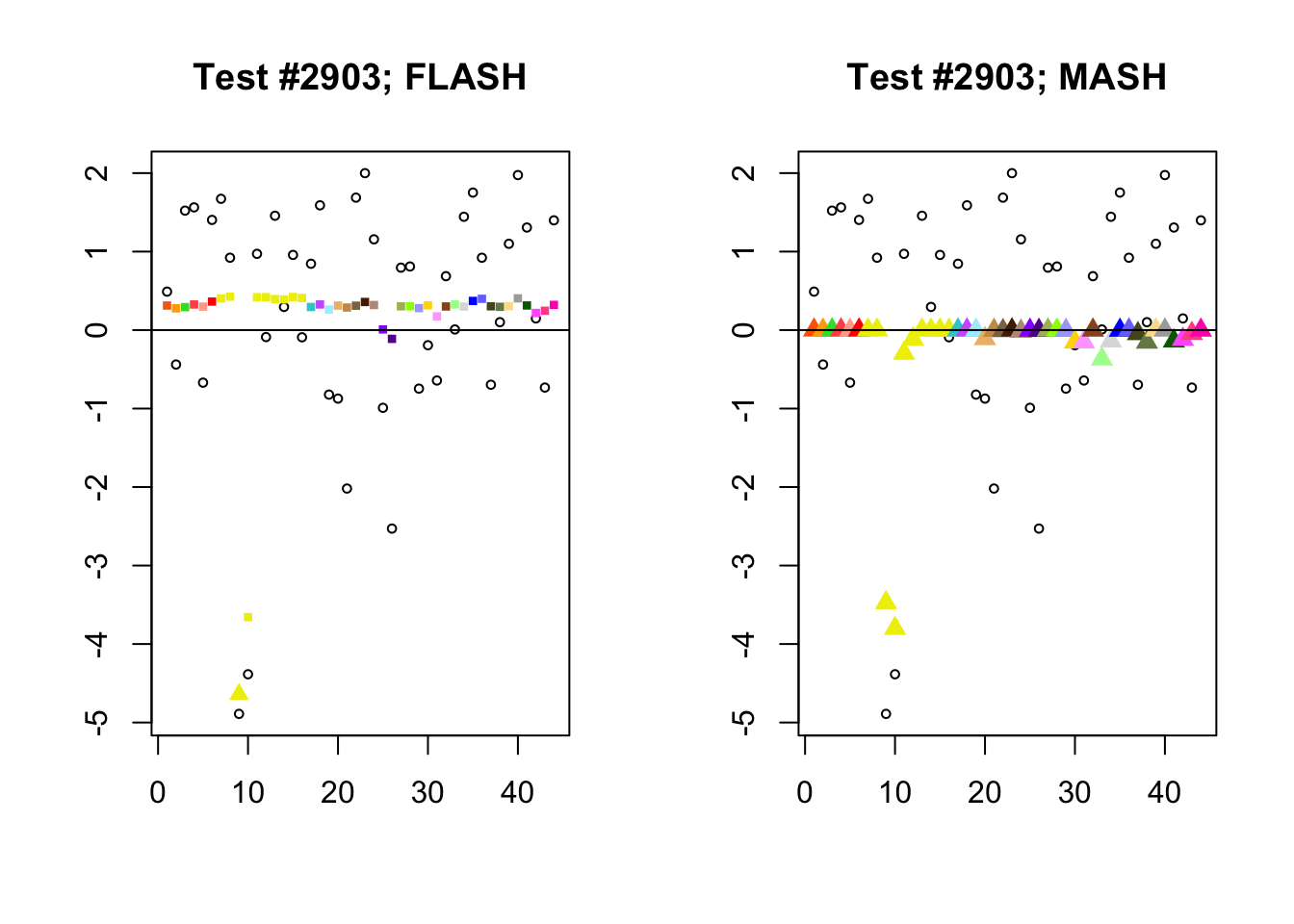
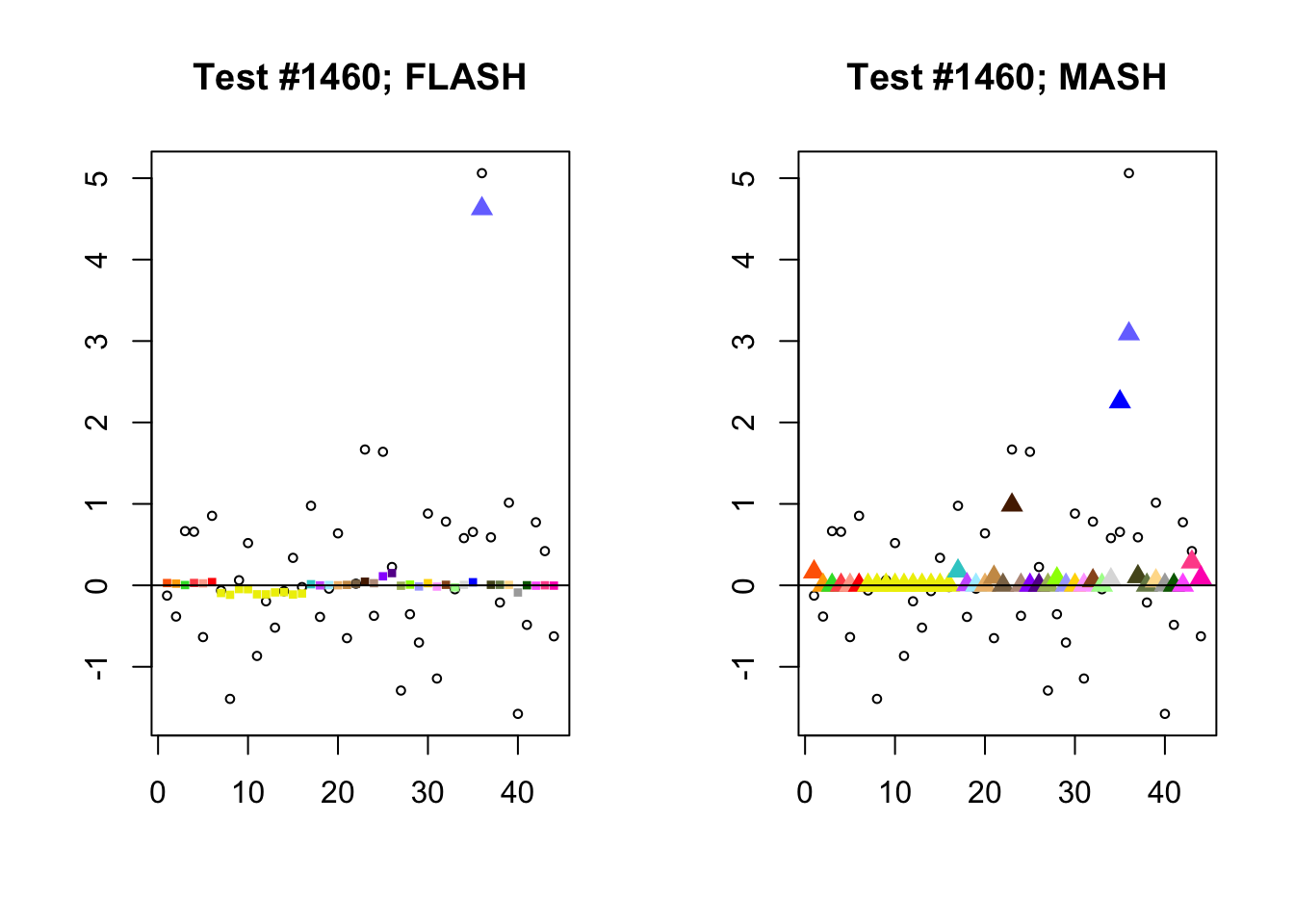
Significant for MASH, not FLASH
In many cases, MASH finds all effects to be significant where FLASH only finds a single unique effect. What happens in these cases, I think, is that MASH puts nearly all posterior weight on a single data-driven covariance matrix. Since these covariance matrices are each rank-one with no entries that are exactly equal to zero, even tiny effects will appear significant (since they are sampled away from zero nearly all of the time).
In contrast, since the FLASH model allows for covariance structures that are linear combinations of these rank-one matrices, the FLASH posteriors are far less likely to follow these patterns of perfect correlation.
par(mfrow=c(1, 2))
mash.not.flash <- c(228, 65, 2903, 1460)
for (n in mash.not.flash) {
plot_test(n, fl_lfsr, fl_pm, "FLASH")
plot_test(n, m_lfsr, m_pm, "MASH")
}
Expand here to see past versions of mash_not_flash-1.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of mash_not_flash-2.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of mash_not_flash-3.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Expand here to see past versions of mash_not_flash-4.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
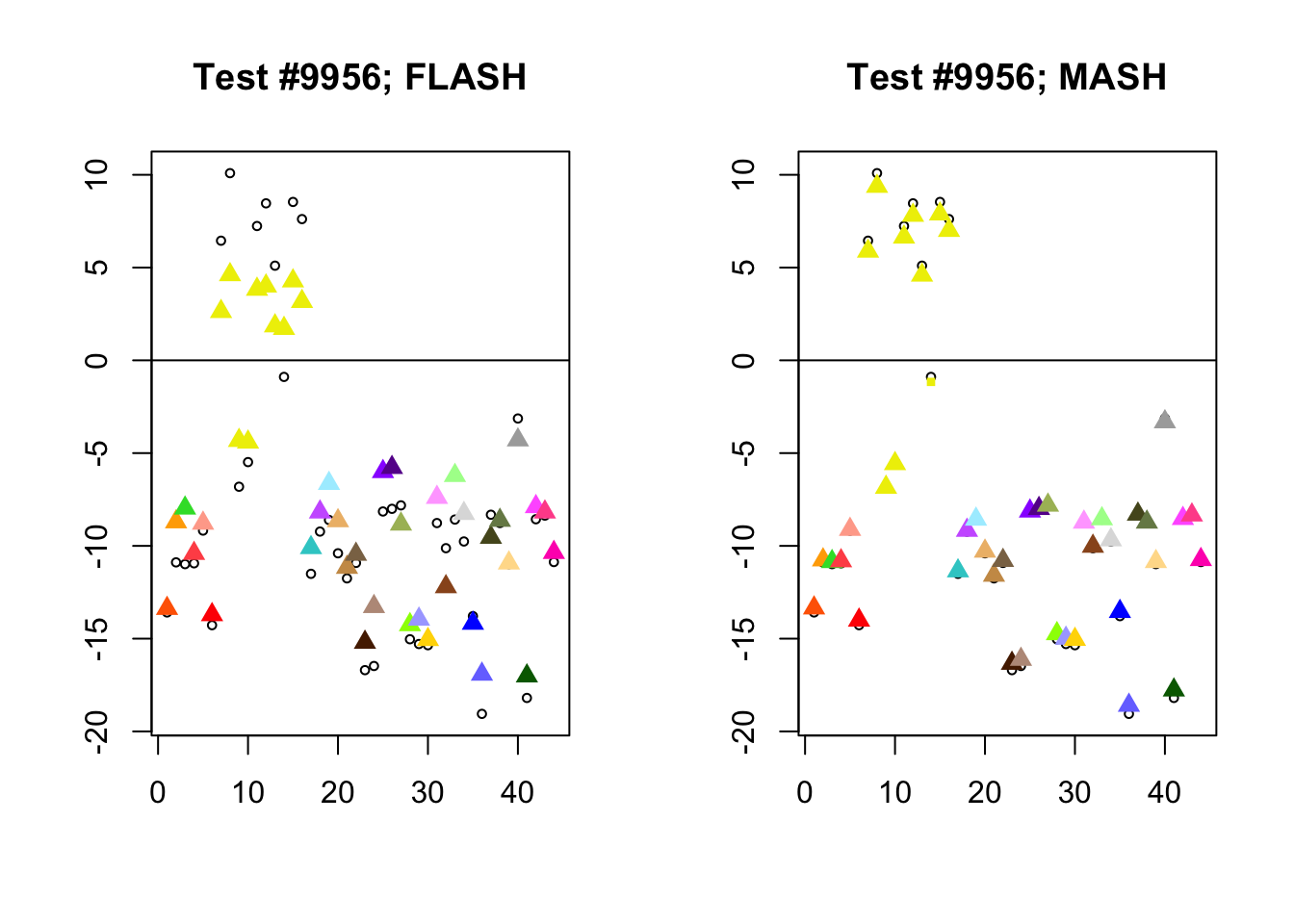
Different posterior means
As discussed here, MASH often does poorly when effect sizes are uniformly large.
par(mfrow=c(1, 2))
diff_pms <- c(9956, 5293, 13168, 3680)
for (n in diff_pms) {
plot_test(n, fl_lfsr, fl_pm, "FLASH")
plot_test(n, m_lfsr, m_pm, "MASH")
}
Expand here to see past versions of diff_pms-1.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
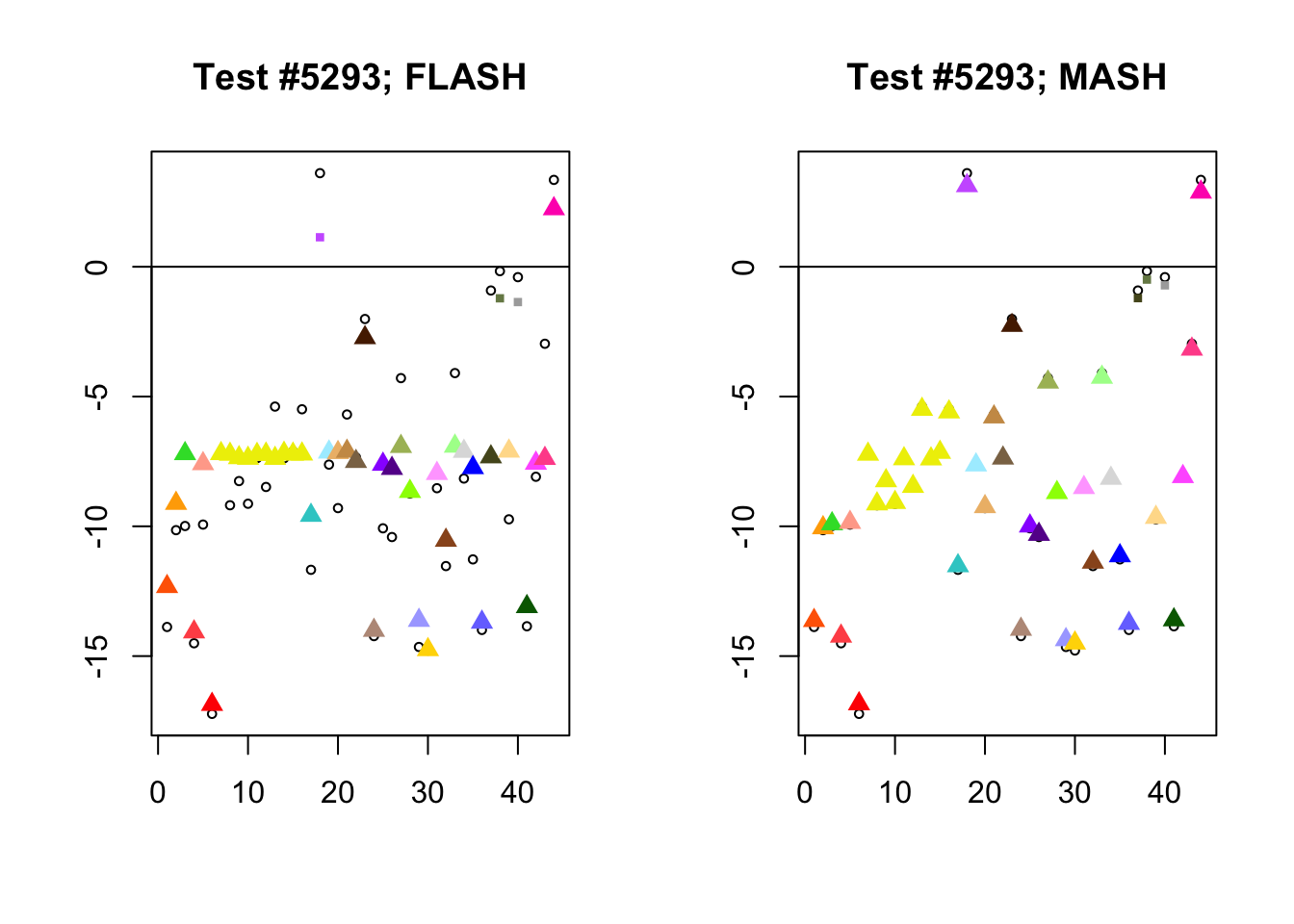
Expand here to see past versions of diff_pms-2.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
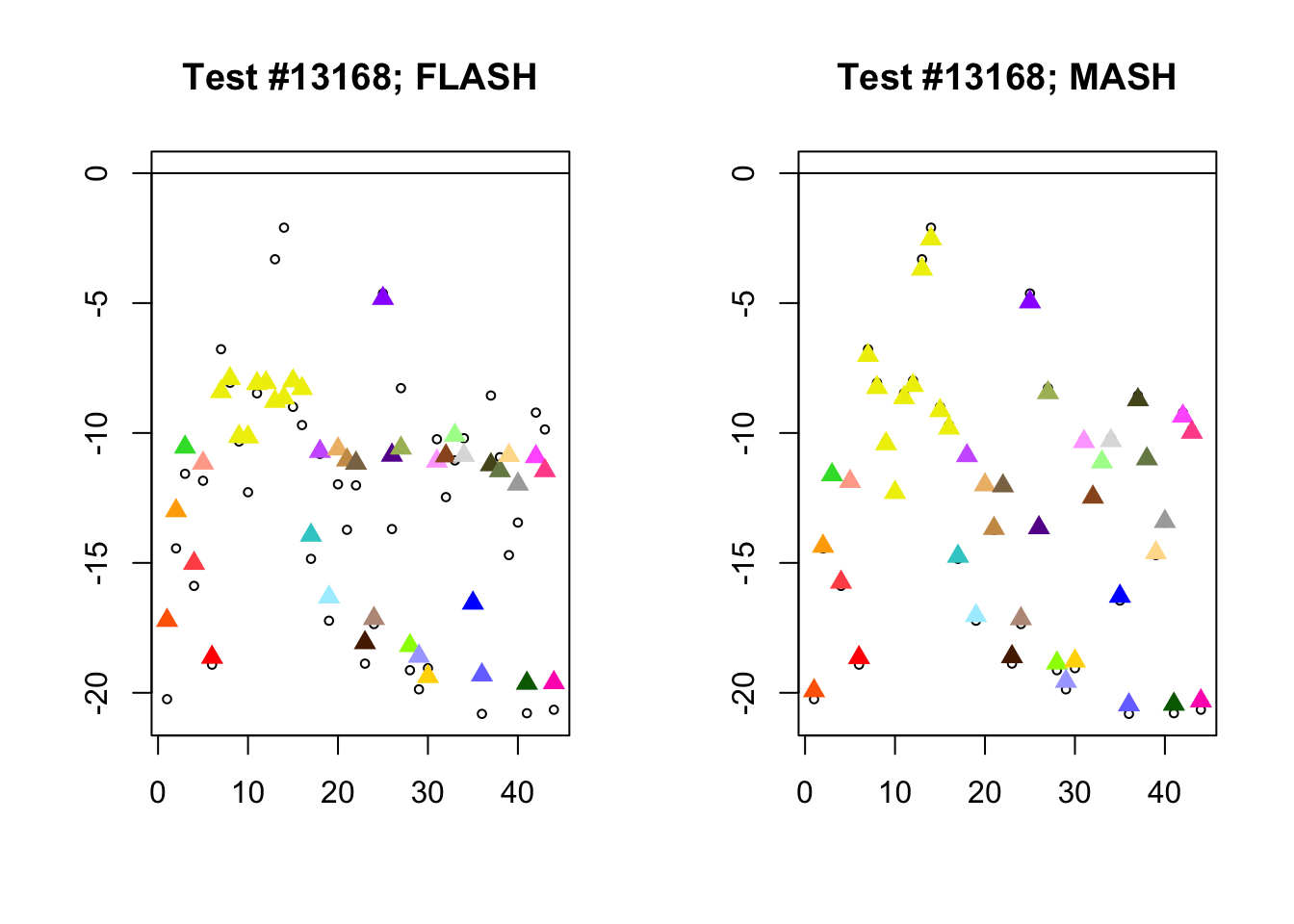
Expand here to see past versions of diff_pms-3.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
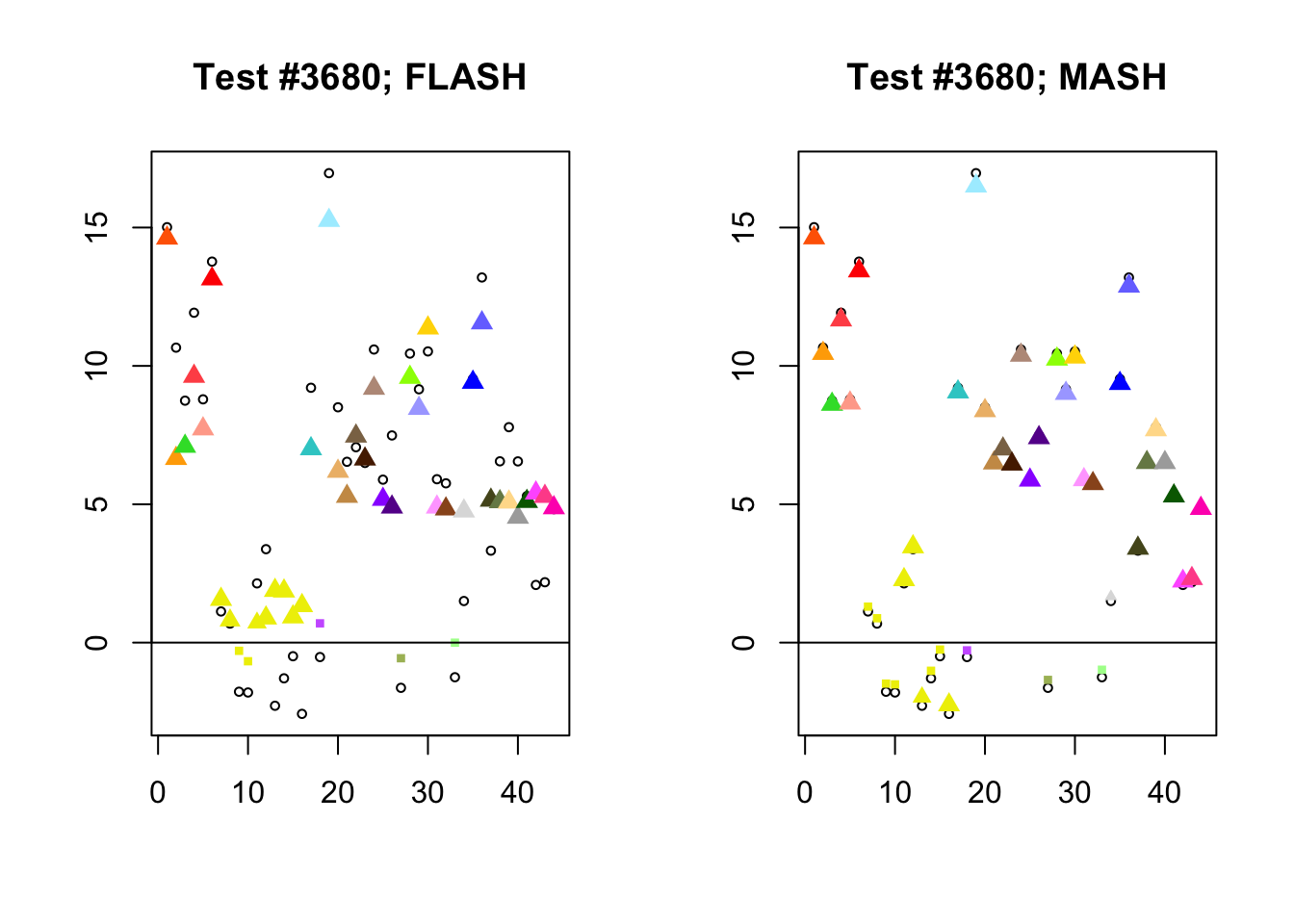
Expand here to see past versions of diff_pms-4.png:
| Version | Author | Date |
|---|---|---|
| 48615e4 | Jason Willwerscheid | 2018-08-15 |
Code
Click “Code” to view the code used to obtain the fits used in this analysis.
# devtools::install_github("stephenslab/flashr")
devtools::load_all("/Users/willwerscheid/GitHub/flashr/")
# devtools::install_github("stephenslab/ebnm")
devtools::load_all("/Users/willwerscheid/GitHub/ebnm/")
library(mashr)
source("./code/utils.R")
gtex <- readRDS(gzcon(url("https://github.com/stephenslab/gtexresults/blob/master/data/MatrixEQTLSumStats.Portable.Z.rds?raw=TRUE")))
strong <- t(gtex$strong.z)
random <- t(gtex$random.z)
strong_data <- flash_set_data(strong, S = 1)
random_data <- flash_set_data(random, S = 1)
# The multi-tissue effects from the previous analysis will serve as
# the data-driven loadings:
nn <- readRDS("./output/MASHvFLASHnn/fl.rds")
multi <- c(2, 5, 6, 8, 11:13, 17, 22:25, 31)
n <- nrow(strong)
dd <- nn$EL[, multi]
dd <- dd / rep(apply(dd, 2, max), each=n) # normalize
## FLASH fit ------------------------------------------------------------
canonical <- cbind(rep(1, n), diag(rep(1, n)))
LL <- cbind(canonical, dd)
fl_random <- flash_add_fixed_loadings(random_data, LL)
system.time(
fl_random <- flash_backfit(random_data,
fl_random,
var_type = "zero",
ebnm_fn = "ebnm_pn",
nullcheck = FALSE)
)
# 26 minutes (62 iterations)
saveRDS(fl_random$gf, "./output/MASHvFLASHgtex3/flgf.rds")
gf <- readRDS("./output/MASHvFLASHgtex3/flgf.rds")
fl_final <- flash_add_fixed_loadings(strong_data, LL)
ebnm_param_f = lapply(gf, function(g) {list(g=g, fixg=TRUE)})
system.time(
fl_final <- flash_backfit(strong_data,
fl_final,
var_type = "zero",
ebnm_fn = "ebnm_pn",
ebnm_param = list(f = ebnm_param_f, l = list()),
nullcheck = FALSE)
)
# 21 minutes (75 iterations)
saveRDS(fl_final, "./output/MASHvFLASHgtex3/fl.rds")
nsamp <- 200
system.time({
sampler <- flash_sampler(strong_data, fl_final, fixed="loadings")
samp <- sampler(200)
fl_lfsr <- flash_lfsr(samp)
})
# 2 minutes
saveRDS(fl_lfsr, "./output/MASHvFLASHgtex3/fllfsr.rds")
# MASH fit --------------------------------------------------------------
strong_data <- mash_set_data(t(strong), Shat = 1)
random_data <- mash_set_data(t(random), Shat = 1)
U.flash <- list()
for (i in 1:ncol(dd)) {
U.flash[[i]] <- outer(dd[, i], dd[, i])
}
U.c <- cov_canonical(random_data)
system.time(m_random <- mash(random_data, Ulist = c(U.c, U.flash)))
# 4 minutes
system.time(
m_final <- mash(strong_data, g=get_fitted_g(m_random), fixg=TRUE)
)
saveRDS(m_final, "./output/MASHvFLASHgtex3/m.rds")
# 19 secondsSession information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] flashr_0.5-14 mashr_0.2-7 ashr_2.2-10
loaded via a namespace (and not attached):
[1] Rcpp_0.12.17 pillar_1.2.1 compiler_3.4.3
[4] git2r_0.21.0 plyr_1.8.4 workflowr_1.0.1
[7] R.methodsS3_1.7.1 R.utils_2.6.0 iterators_1.0.9
[10] tools_3.4.3 testthat_2.0.0 digest_0.6.15
[13] tibble_1.4.2 gtable_0.2.0 evaluate_0.10.1
[16] memoise_1.1.0 lattice_0.20-35 rlang_0.2.0
[19] Matrix_1.2-12 foreach_1.4.4 commonmark_1.4
[22] yaml_2.1.17 parallel_3.4.3 ebnm_0.1-12
[25] mvtnorm_1.0-7 xml2_1.2.0 withr_2.1.1.9000
[28] stringr_1.3.0 knitr_1.20 roxygen2_6.0.1.9000
[31] devtools_1.13.4 rprojroot_1.3-2 grid_3.4.3
[34] R6_2.2.2 rmarkdown_1.8 rmeta_3.0
[37] ggplot2_2.2.1 magrittr_1.5 whisker_0.3-2
[40] scales_0.5.0 backports_1.1.2 codetools_0.2-15
[43] htmltools_0.3.6 MASS_7.3-48 assertthat_0.2.0
[46] softImpute_1.4 colorspace_1.3-2 stringi_1.1.6
[49] lazyeval_0.2.1 munsell_0.4.3 doParallel_1.0.11
[52] pscl_1.5.2 truncnorm_1.0-8 SQUAREM_2017.10-1
[55] R.oo_1.21.0 This reproducible R Markdown analysis was created with workflowr 1.0.1